Parallel Workflows on Kubernetes
Learn how parallel workflows running in distributed systems on Kubernetes can be used to solve two problems.
Join the DZone community and get the full member experience.
Join For FreeApplications are now increasingly distributed, running on multiple machines and accessed by multiple users from all over the world. By bundling the application code, the application runtime, and the libraries, containers, and container orchestrators, we have addressed many of the challenges of building distributed systems. With container runtimes like Docker, we can deploy our applications on different environments. And with container orchestration tools like Kubernetes, we can scale our applications. We frequently have to break our distributed application into a collection of multiple containers running on different machines. This requires us to coordinate execution and facilitate communication among different containers. A scenario where this situation is encountered is when we compose a workflow using multiple containers. In this article, we will learn how to build such workflows using a container workflow engine, Argo, for Kubernetes. We will develop workflows for the following two examples of so-called "embarrassingly parallel" problems for which Kubernetes is the ideal scaling platform.
N-Queens using genetic algorithms
Distributed search
N-Queens Using Genetic Algorithms
The N-Queens problem is to place N queens on an NxN chessboard so that no two attack. Since each queen must be on a different row and column, we can assume that queen i is placed in i-th column. All solutions to the N-Queens problem can, therefore, be represented as N-tuples (q1, q2, …, qN) that are permutations of an N-tuple (1, 2, 3, …, N). The position of a number in the tuple represents the queen's column position, while its value represents the queen's row position. The complexity of the search is N*(N-1)* ... *1 = N!.
Genetic Algorithms
Genetic algorithms can be used to search for a permutation of N non-attacking queens. The pseudo-code shown below is a simple genetic algorithm to search for a solution.
create initial population
evaluate initial population
while not done
select 3 individuals
run mutation operator
evaluate offspring
if solution found, set done = true
endThe choice of initial population, random here, determines how fast the solution is found. There are different ways to parallelize this algorithm to speed it up. One of the simplest ways is to run this same computation on multiple workers with a different randomly-selected initial population. Once any worker has exited with success, no other worker should still be doing any work or writing any output. They should all exit as soon as one of the workers finds the solution.
Argo Workflow
Let us create a workflow to be run on Kubernetes that runs this genetic algorithm along with other subsequent processing tasks. A Kubernetes Job with a specified number of completions and parallelism is used for parallel execution of pods. We create a Job object with 5 completions and 5 parallelism that will launch 5 pods in parallel to search for a solution. A Redis queue service is used for pubsub. The pod that finishes first will publish a message on a channel on the Redis server. All workers are subscribed to the channel. On receiving "finished" message, all other workers stop the search and exit.
The Docker image containing the Python implementation of the genetic algorithm as described above can be pulled from the DockerHub. This image requires a Redis service to be running on the Kubernetes cluster. Refer to the Kubernetes documentation on how to create and configure a Redis service on Kubernetes.
> docker pull randhirkumars/n-queens-genetic-redisThe workflow then consists of the following steps:
Create a parallelized Kubernetes Job which launches 5 parallel workers. Once any pod has exited with success, no other pod will be doing any work. Return the job name and job uid as output parameters.
Using the uid of the job, query any of its associated pods and print the result to the stdout.
Delete the job using the job name.
The Argo workflow YAML with the above steps is shown below:
apiVersion: argoproj.io/v1alpha1
kind: Workflow
metadata:
generateName: nqueens-
spec:
entrypoint: nqueens-job
templates:
- name: nqueens-job
steps:
- - name: nqueens-genetic-job
template: nqueens-genetic-job
- - name: print-solution
template: print-solution
arguments:
parameters:
- name: job-uid
value: '{{steps.nqueens-genetic-job.outputs.parameters.job-uid}}'
- - name: delete-job
template: delete-job
arguments:
parameters:
- name: job-name
value: '{{steps.nqueens-genetic-job.outputs.parameters.job-name}}'
- name: nqueens-genetic-job
resource:
action: create
successCondition: status.succeeded > 1
failureCondition: status.failed > 0
manifest: |
apiVersion: batch/v1
kind: Job
metadata:
namespace: default
name: nqueens
labels:
job: n-queens
spec:
parallelism: 5
completions: 5
template:
metadata:
labels:
job: n-queens
spec:
containers:
- name: worker
image: randhirkumars/n-queens-genetic-redis
restartPolicy: Never
outputs:
parameters:
- name: job-name
valueFrom:
jsonPath: '{.metadata.name}'
- name: job-uid
valueFrom:
jsonPath: '{.metadata.uid}'
- name: print-solution
inputs:
parameters:
- name: job-uid
container:
image: argoproj/argoexec:latest
command: [sh, -c]
args: ["
for i in `kubectl get pods -l controller-uid={{inputs.parameters.job-uid}} -o name`; do
kubectl logs $i;
done
"]
- name: delete-job
inputs:
parameters:
- name: job-name
resource:
action: delete
manifest: |
apiVersion: batch/v1
kind: Job
metadata:
name: {{inputs.parameters.job-name}}Notice that each step in the workflow is a Docker container. The first step uses a Docker image that we have built — randhirkumars/n-queens-genetic-redis — and the second step uses a pre-built image from DockerHub — argoproj/argoexec:latest. Submit the workflow using the Argo command line.
> argo submit nqueens.yamlThis launches the workflow and this can be visualized as a graph in Argo UI.
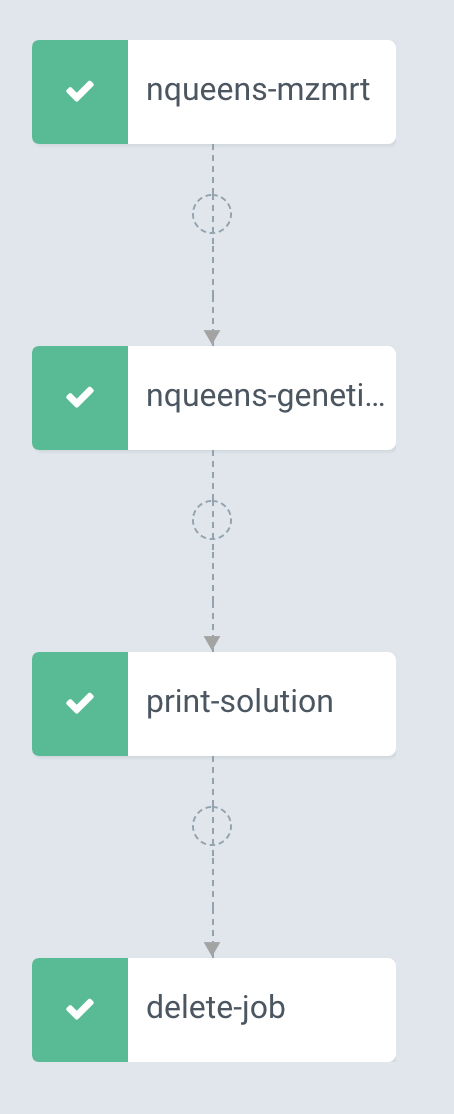
Once the workflow has run to completion, the solution is printed out to the console of the pod associated with the Job. A solution for a 14x14 chessboard is shown below.

Distributed Search
In the N-Queens problem, we learned how to create a workflow that launched parallel jobs on Kubernetes. The parallel jobs used an external Redis queue service for coordination. In this section, we look at another pattern for parallel workflow — distributed document search using the scatter/gather pattern. Here, the task is to search different words across a large database of documents for all documents that contain those words. To parallelize the task, we will scatter the different term requests across nodes in the cluster. All the nodes in the cluster have access to a shared volume hosting the documents to be searched. Then we gather all the responses from worker nodes into a single response. This task can be implemented in a workflow with the following steps.
When a request comes into search, parse the request and split the search string into words.
Loop through the words and farm out a leaf pod to search for each word.
Each of the pods returns a list of documents that match one of the words. Collate the search results and print the list of documents.
Argo Workflow
The Argo workflow YAML with the above steps is shown below. Here, we store the documents to be searched and the results of the search on persistent volumes mounted on the pods.
apiVersion: argoproj.io/v1alpha1
kind: Workflow
metadata:
generateName: dgrep-
spec:
entrypoint: dgrep
volumeClaimTemplates:
- metadata:
name: workdir
spec:
accessModes: [ "ReadWriteOnce" ]
resources:
requests:
storage: 1Gi
arguments:
parameters:
- name: searchstring
value: hello world
templates:
- name: dgrep
steps:
- - name: data
template: data
- - name: generate
template: generate
- - name: search
template: search
arguments:
parameters:
- name: words
value: "{{item}}"
withParam: "{{steps.generate.outputs.result}}"
- - name: collate
template: collate
- name: data
container:
image: alpine
command: [sh, -c]
args: ["touch /mnt/data/file1; touch /mnt/data/file2; echo -n hello > /mnt/data/file1; echo -n world > /mnt/data/file2"]
volumeMounts:
- name: workdir
mountPath: /mnt/data
- name: generate
script:
image: python:alpine3.6
command: [python]
source: |
import json
import sys
json.dump([w for w in "{{workflow.parameters.searchstring}}".split()], sys.stdout)
- name: search
inputs:
parameters:
- name: words
container:
image: alpine
command: [sh, -c]
args: ["grep -rl {{inputs.parameters.words}} /mnt/data | awk -F/ '{ print $NF }' >> /mnt/data/output"]
volumeMounts:
- name: workdir
mountPath: /mnt/data
- name: collate
container:
image: alpine
command: [sh, -c]
args: ["cat /mnt/data/output | sort | uniq"]
volumeMounts:
- name: workdir
mountPath: /mnt/data
Here we are using the vanilla Docker image alpine for all the steps. The workflow, when submitted using the Argo command line, can be visualized on the Argo UI. The search string consists of two words, "hello" and "world." In the response, two pods are created in parallel, each searching for a word. The individual search results from the pods are collated in the final step to print the list of documents.

Conclusion
We have illustrated how to create parallel workflows on Kubernetes with the help of two examples. We used Argo to create workflows that can be specified as a directed acyclic graph (DAG). Argo allows us to define a container-native workflow on Kubernetes where each step in the workflow is a Docker container. Kubernetes is ideal for running parallel workflows and Argo reduces the complexity of designing such workflows.
Opinions expressed by DZone contributors are their own.

Comments