Use pgvector With PostgreSQL To Improve LLM Accuracy and Performance
This article shows how to improve LLM accuracy and performance with pgvector and PostgreSQL. It explores embeddings and why the role of pgvector is crucial.
Join the DZone community and get the full member experience.
Join For FreeIf you’re not yet familiar with the open-source pgvector extension for PostgreSQL, now’s the time to do so. The tool is extremely helpful for searching text data fast without needing a specialized database to store embeddings.
Embeddings represent word similarity and are stored as vectors (a list of numbers). For example, the words “tree” and “bush” are related more closely than “tree” and “automobile.” The open-source pgvector tool makes it possible to search for closely related vectors and find text with the same semantic meaning. This is a major advance for text-based data, and an especially valuable tool for building Large Language Models (LLMs)... and who isn’t right now?
By turning PostgreSQL into a high-performance vector store with distance-based embedding search capabilities, pgvector allows users to explore vast textual data easily. This also enables exact nearest neighbor search and approximate nearest neighbor search using L2 (or Euclidian) distance, inner product, and cosine distance. Cosine distance is recommended by OpenAI for capturing semantic similarities efficiently.
Using Embeddings in Retrieval Augmented Generation (RAG) and LLMs
Embeddings can play a valuable role in the Retrieval Augmented Generation (RAG) process, which is used to fine-tune LLMs on new knowledge. The process includes retrieving relevant information from an external source, transforming it into an LLM digestible format, and then feeding it to the LLM to generate text output.
Let’s put an example to it. Searching documentation for answers to technical problems is something I’d bet anyone here has wasted countless hours on. For this example below, using documentation as the source, you can generate embeddings to store in PostgreSQL. When a user queries that documentation, the embeddings make it possible to represent the words in a query as vector numbers, perform a similarity search, and retrieve relevant pieces of the documentation from the database. The user’s query and retrieved documentation are both passed to the LLM, which accurately delivers relevant documentation and sources that answer the query.
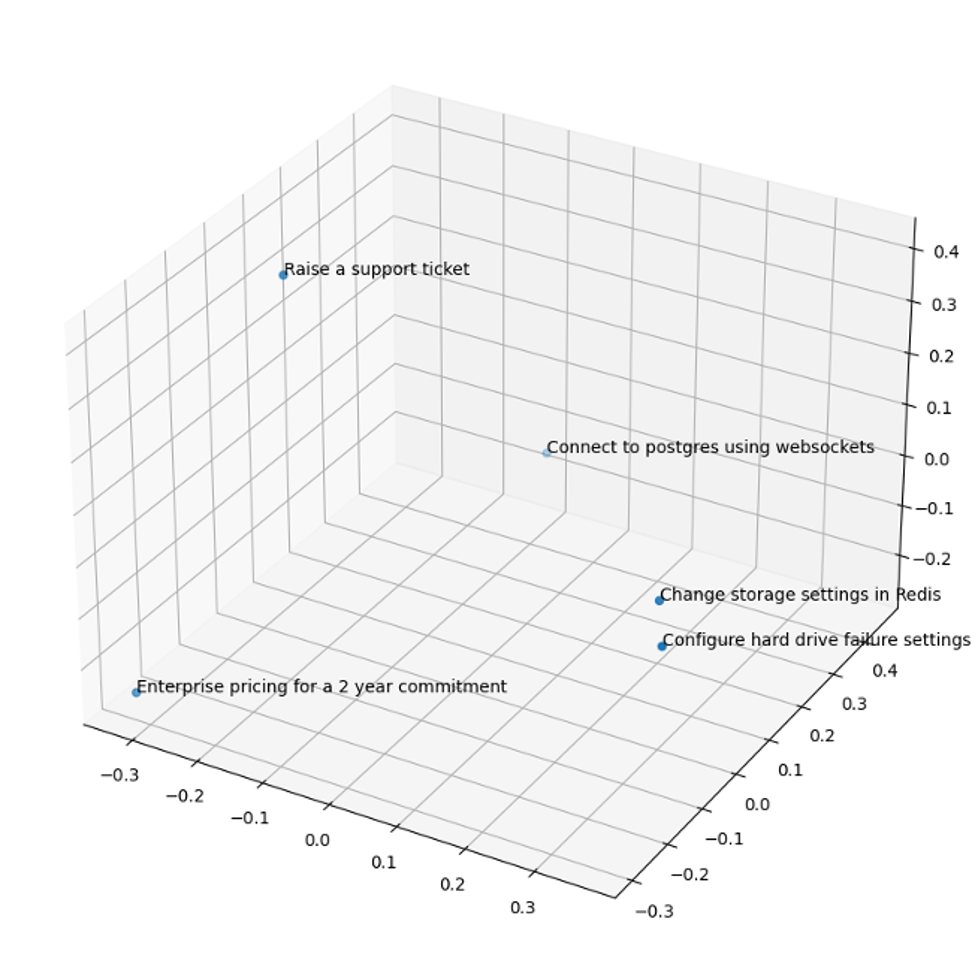
We tested out pgvector and embeddings using our own documentation at Instaclustr. Here are some example user search phrases to demonstrate how embeddings will plot them relative to one another:
- “Configure hard drive failure setting in Apache Cassandra”
- “Change storage settings in Redis”
- “Enterprise pricing for a 2-year commitment”
- “Raise a support ticket”
- “Connect to PostgreSQL using WebSockets”
Embeddings plot the first two phases nearest each other, even though they include none of the same words.
The LLM Context Window
Each LLM has a context window: the number of tokens it can process at once. This can be a challenge, in that models with a limited context window can falter with large inputs, but models trained with large context windows (100,000 tokens, or enough to use a full book in a prompt) suffer from latency and must store that full context in memory. The goal is to use the smallest possible context window that generates useful answers. Embeddings help by making it possible to provide the LLM with only data recognized as relevant so that even an LLM with a tight context window isn’t overwhelmed.
Feeding the Embedding Model With LangChain
The model that generates embeddings — OpenAI’s text-embedding-ada-002 — has a context window of its own. That makes it essential to break documentation into chunks so this embedding model can digest more easily.
The LangChain Python framework offers a solution. An LLM able to answer documentation queries needs these tasks completed first:
- Document loading: LangChain makes it simple to scrape documentation pages, with the ability to load diverse document formats from a range of locations.
- Document transformation: Segmenting large documents into smaller digestible chunks enables retrieval of pertinent document sections.
- Embedding generation: Calculate embeddings for the chunked documentation using OpenAI’s embedding model.
- Data storing: Store embeddings and original content in PostgreSQL.
This process yields the semantic index of documentation we’re after.
An Example User Query Workflow
Now consider this sample workflow for a user query (sticking with our documentation as the example tested). First, a user submits the question: “How do I create a Redis cluster using Terraform?” OpenAI’s embeddings API calculates the question’s embeddings. The system then queries the semantic index in PostgreSQL using cosine similarity, asking for the original content closest to the embeddings of the user’s question. Finally, the system grabs the original content returned in the vector search, concatenates it together, and includes it in a specially crafted prompt with the user’s original question.
Implementing pgvector and a User Interface
Now let’s see how we put pgvector into action. First, we enabled the pgvector extension in our PostgreSQL database, and created a table for storing all documents and their embeddings:
CREATE EXTENSION vector;
CREATE TABLE insta_documentation (id bigserial PRIMARY KEY, title, content, url, embedding vector(3));The following Python code scrapes the documentation, uses Beautiful Soup to extract main text parts such as title and content, and stores them and the URL in the PostgreSQL table:
urls = [...]
def init_connection():
return psycopg2.connect(**st.secrets["postgres"])
def extract_info(url):
hdr = {'User-Agent': 'Mozilla/5.0'}
req = Request(url,headers=hdr)
response = urlopen(req)
soup = BeautifulSoup(response, 'html.parser')
title = soup.find('title').text
middle_section = soup.find('div', class_='documentation-middle').contents
# middle section consists of header, content and instaclustr banner and back and forth links - we want only the first two
content = str(middle_section[0]) + str(middle_section[1])
return title, content, url
conn = init_connection()
cursor = conn.cursor()
for url in urls:
page_content = extract_info(url)
postgres_insert_query = """ INSERT INTO insta_documentation (title, content, url) VALUES (%s, %s, %s)"""
cursor.execute(postgres_insert_query, page_content)
conn.commit()
if conn:
cursor.close()
conn.close()Next, we loaded the documentation pages from the database, divided them into chunks, and created and stored the crucial embeddings.
def init_connection():
return psycopg2.connect(**st.secrets["postgres"])
conn = init_connection()
cursor = conn.cursor()
# Define and execute query to the insta_documentation table, limiting to 10 results for testing (creating embeddings through the OpenAI API can get costly when dealing with a huge amount of data)
postgres_query = """ SELECT title, content, url FROM insta_documentation LIMIT 10"""
cursor.execute(postgres_query)
results = cursor.fetchall()
conn.commit()
# Load results into pandas DataFrame for easier manipulation
df = pd.DataFrame(results, columns=['title', 'content', 'url'])
# Break down content text which exceed max input token limit into smaller chunk documents
# Define text splitter
html_splitter = RecursiveCharacterTextSplitter.from_language(language=Language.HTML, chunk_size=1000, chunk_overlap=100)
# We need to initialize our embeddings model
embeddings = OpenAIEmbeddings(model="text-embedding-ada-002")
docs = []
for i in range(len(df.index)):
# Create document with metadata for each content chunk
docs = docs + html_splitter.create_documents([df['content'][i]], metadatas=[{"title": df['title'][i], "url": df['url'][i]}])
# Create pgvector dataset
db = Pgvector.from_documents(
embedding=embeddings,
documents=docs,
collection_name=COLLECTION_NAME,
connection_string=CONNECTION_STRING,
distance_strategy=DistanceStrategy.COSINE,
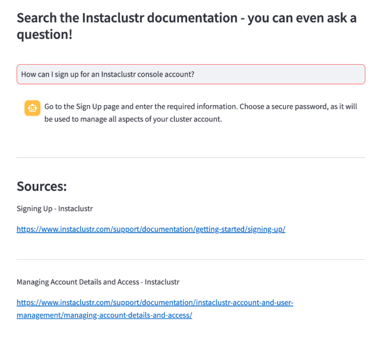
)Lastly, the retriever found the correct information to answer a given query. In our test example, we searched our documentation to learn how to sign up for an account:
query = st.text_input('Your question', placeholder='How can I sign up for an Instaclustr console account?')
retriever = store.as_retriever(search_kwargs={"k": 3})
qa = RetrievalQA.from_chain_type(
llm=OpenAI(),
chain_type="stuff",
retriever=retriever,
return_source_documents=True,
verbose=True,
)
result = qa({"query": query})
source_documents = result["source_documents"]
document_page_content = [document.page_content for document in source_documents]
document_metadata = [document.metadata for document in source_documents]Using Streamlit, a powerful tool for building interactive Python interfaces, we built this interface to test the system and view the successful query results:
Data Retrieval With Transformative Efficiency
Harnessing PostgreSQL and the open-source pgvector project empowers users to leverage natural language queries to answer questions immediately, with no need to comb through irrelevant data. The result: super accurate, performant, and efficient LLMs, groundbreaking textual capabilities, and meaningful time saved!
Opinions expressed by DZone contributors are their own.

Comments