A Clinical Decision Support System Built With a Knowledge Graph
Using Open Source technologies to Improve Doctor-Patient Engagement
Join the DZone community and get the full member experience.
Join For FreeDebrief from a Grakn Community talk — featuring Alessia Basadonne, executive PHD candidate from University of Pavia and Medas Italy. This talk was delivered live at Grakn Cosmos 2020 in London, UK.
“From when I was very very little, I always dreamed of developing crazy ideas and making them a reality.”
Alessia’s current work is in developing a Clinical Decision Support System (CDSS). This isn’t a new concept as she highlights, but one with a lot of opportunity for improvements and developments. So…
What’s the Problem these Systems Help to Solve?
How long does a patient spend with a doctor, on average?
With the exception of countries that self-report, the average was around 9 minutes per encounter. Alessia notes that after speaking with doctors and nurses, the reality is closer to 5 minutes. Now imagine what needs to be accomplished by the doctor in those 5 minutes:
- Review all existing patient data and history
- Recall all of their personal medical experience
- Provide diagnosis, treatment or behaviour recommendation to the patient
This is a tall order, one that can lead to missing information, miss-diagnosis or at worst, a recommendation of treatment that causes adverse effects on the patient.
What’s the Solution?
A Clinical Decision Support System provides the doctor with a tool that eases their work, increases the value of the short time spent in the room with an individual, and ensures that the full context is available to the doctor.
Let’s look at what using a tool like this might look like in the field:
- The doctor collects all the patient information from heterogeneous data sources and inputs it into the system
- The doctor then consults the CDSS during an appointment
- The CDSS compares the patient information with the guidelines and returns a recommendation
- These recommendations need to be specific for both the patient and the guidelines of the region, country or hospital to ensure that recommendations are of high value — obvious recommendations are not valuable here
- Finally the doctor provides an answer to the patient with the full context from the doctor’s experience and the CDSS’ set of medical information
Alessia notes that ultimately the recommendation provided by the CDSS must be specific to the guidelines context of the current landscape as well as the situation occurring with a particular patient. The guidelines mentioned above are just a part of the complexity involved in working with medical data. Guidelines are the collective sum of the most up to date medical literature in a specific field.
Expert System as a CDSS
A system that manages and reasons over such a breadth of information such as: regional regulatory documents, situational recommendations, and patient data; starts to sound like an expert system.
Going strictly by the book, an expert system is defined as…
“A computer system that emulates, or acts in all respects, with the decision making capabilities of a human expert.”
Professor Edward Feigenbaum — Stanford University
What are the Components of an Expert System?
Knowledge Base — contains facts, in this case medical facts, to establish ground truth
Reasoning engine — reasoning across the knowledge base to provide recommendations based on scenarios
Interface — a user-facing platform to enable access and querying
What Work has Been Done Already?
in bringing expert systems to the medical decision domain…
As a concept this has been around a while. In the medical domain however, there’s much to be desired. In the 1970s there were a few expert systems built as clinical decision support systems, such as De Dombal’s original CDSS in 1970, focused on acute abdominal pain. INTERNIST, built in 1974, was a general internal medicine support system. While De Dombal used naive bayes as a guide for the reasoning, MYCIN was the first rule based CDSS built in 1976.
It is remarkable that between MYCIN in 1976 and the turn of the century, there was almost no development done to improve or innovate in this space. In the last 20 years interest has grown and a new found effort has been placed into empowering medical recommendations with technology. Today, we have just one CDSS in production. However, in the medical field there is little to no examples or projects using a knowledge graph for such systems.
How are Rules Used in Past and Current Systems?
Take a look at the below production rule from a 1970s system — you’ll notice that the rule is made up of a series of AND statements in the if clause, and concludes with THEN diagnosis =. We can imagine that using these rules can get fairly complex and messy. The CDSS currently in production in Italy, Alessia notes, is using rules like this. Leaving a lot of room for development.

Alessia, keying in on this opportunity sought to use graph technology as a way to improve both the method of developing a CDSS as well as the value it brings to doctors and their patients.
Introducing SOPHIA

SOPHIA is a CDSS built with a text-mined knowledge graph. Patient data, mined from unstructured medical records text, is matched with the text of one or multiple guidelines. The guidelines used are aligned-to by the development team and the hospital facility. This is necessary, as guidelines can be specific to hospitals, regions, and countries; as they can be slightly different, you need to have alignment on what guidelines are to be used.

Here we get a look at the data flow within SOPHIA. As Alessia is developing this CDSS in partnership with Medas, their medical technology products are used for passing patient data into SOPHIA. The three products used are:
- MefoRT — an electronic record in radiation therapy
- VPPower —a reporting system in OBGYN
- MeView — a patient portal
Each of these is able to produce a free text report which is then inserted into SOPHIA, making up the NLP pipeline for the project.
These reports are then combined with the NLP output of the medical guidelines. In the future, Alessia would like to incorporate existing, publicly available datasets like: Pubmed or Wolters Kluwer. These would be used to match the bibliographic data from the guidelines to these datasets.
For the end user SOPHIA outputs three types of data:
- Information —a match is found between the patient data and the guidelines
- Alert — when the recommendation suggests a behaviour
- Tailored Recommendation
What’s the Stack Inside SOPHIA?
Traditionally, or rather for most engineers looking to build such an expert system using a knowledge graph and some NLP output, would require a host of technologies and languages from the semantic web. Alessia didn’t go down that path — choosing to use Grakn as the database, and its query language Graql, for its simplicity and expressiveness. Before we get there however, let’s take a look at the pieces needed to build a clinical decision support system or personal care agent.
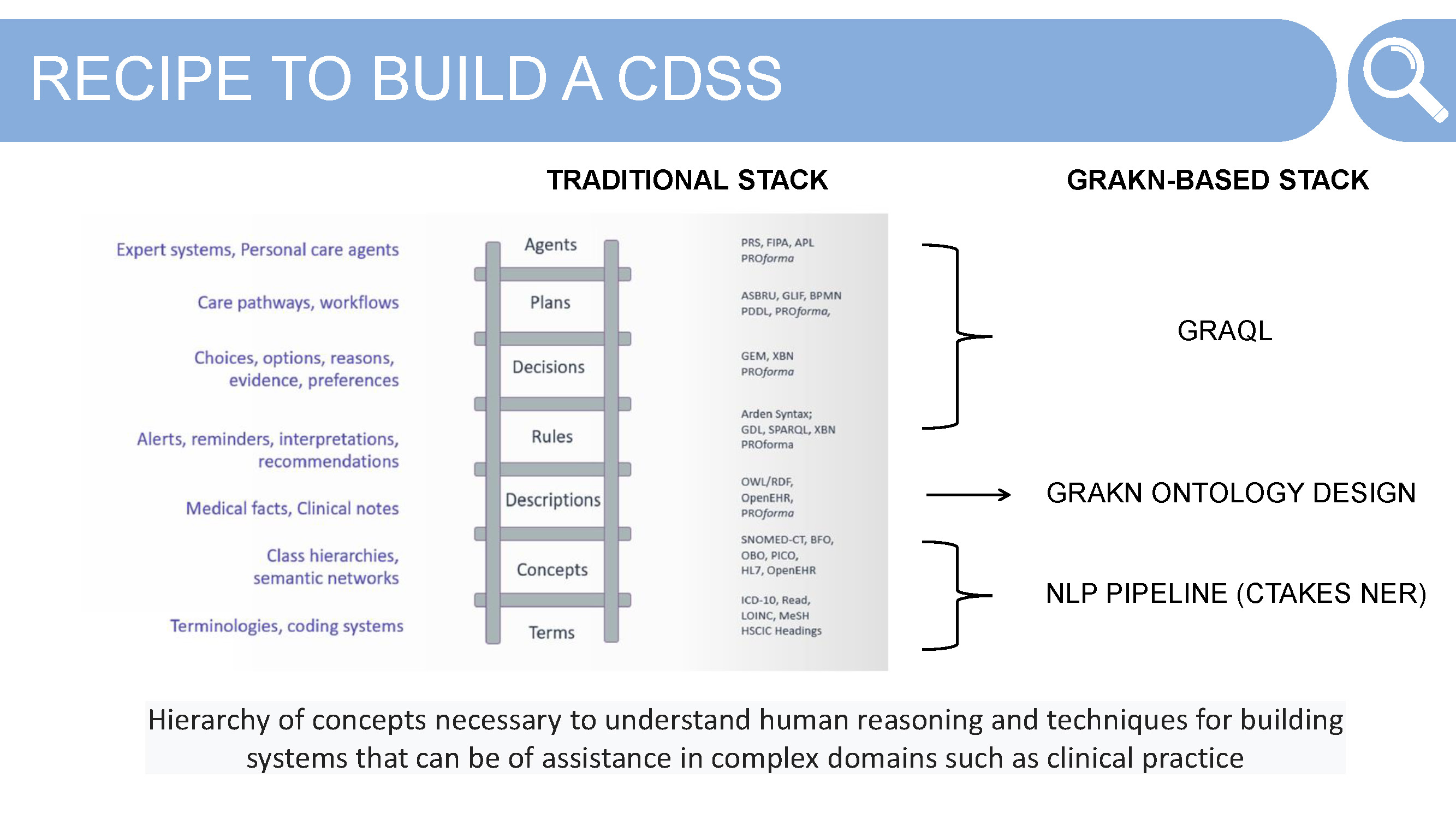
Identify and Align to Terms — terminology can cause problems in a system that uses multiple sources of data in the same domain, a headache and a migraine for example. Alignment is necessary to before building the Grakn schema
Define Concepts — take these aligned-to terms and solidify the hierarchy of concepts that are necessary for the system. This often takes some thinking about what questions the end user will end up asking of the system, as well as how the concepts relate to each other in their real world context.
These terms and concept hierarchies are derived from an NLP pipeline — using the tool CTAKES — and its named entity recognition (NER) tool. The defined ontology is the knowledge representation work required to create the structure to map against the NLP output from the clinical guidelines and patient data from the Medas products. Here Alessia makes use of Grakn schema.
Descriptions, Rules, Decisions, Plans and Agent — these components are where the “fun” starts. Now that we have some NLP output and a concept level Grakn schema, how do we go about matching — finding relations between — the medical guidelines and patient data?
Your first thought might be to use machine learning or do some deep learning here. As Alessia realised, these weren’t necessary when using the Grakn database. Using Graql, Grakn’s query language, was simple and effective for creating these connections without needing to use machine learning or deep learning techniques.
Building SOPHIA
Now that we have a bit of context of where these systems have come from, and the components needed; let’s go through a step by step guide to building a Knowledge Graph based Clinical Decision Support System.
1. Identify Input Data to be Used
The first step is to crystalise what data you plan on migrating into Grakn. These can be structured and unstructured data, output from an NLP pipeline, patient demographic data, etc. Alessia uses a combination of all these:
- Unstructured text from Medical Reports — from the medical device or product
- Patient demographic information such as age, gender, etc.
- NLP output from medical guidelines text
- Future additions would be publicly available datasets such as Pubmed
2. Identify Text Mining Tool
The next step is Alessia’s favourite. It’s important to assess the NLP tools available through the use case you are working in. Tools like Stanford Core NLP and Spacy are useful for general domain use cases. Stanford Core NLP requires a custom named entity recognition model, which for the medical domain would require a lot of work to make effective.
The two other tools assessed for SOPHIA were Apache cTAKES and Amazon Comprehend Medical. The latter does some impressive things but does come with a financial investment. Apache cTAKES is an open source system widely used in the medical community and one that has seen its popularity grow in the wake of the COVID-19 pandemic. ApacheCon this past year dedicated an entire track to the tool. The training work that makes Core NLP and Spacy too resource intensive to set up, isn’t required for both of these systems, making them ideal solutions for the medical and life sciences domains.
Ultimately, Alessia chose Apache cTAKES for the NLP work within SOPHIA. However, Alessia did note that the desktop application wasn’t helpful given she works in web apps. We can see below a selection of the key pipelines that the tool can handle:
- Boundary detection — able to identify the sentences inside of text
- Tokenisation — able to extract single tokens from a sentence
- Normalisation (Lemma) — able to understand similar or like words
- Part-of-speech — able to extract parts of speech within a sentence
- Shallow parsing — able to take those parts and link them with higher order groups (noun groups or phrases, verb groups, etc)
- Entity recognition — able to extract named entities in unstructured text and classify them into defined categories, this is one of the most well loved components
Apache cTAKES is plug and play from the download, which as we mentioned, demonstrates the benefits of not needing to do the domain specific model training. However, as you might notice, the output provided by the system isn’t useable for any development work. Requiring Alessia to do some difficult transformation of the output data to enable usage in development.
3. Mine Text
Having annotated the entities and dropped some free text — italian medical guidelines in Alessia’s case — into Apache cTAKES, the output is then transformed into a low level JSON. This output is then able to be utilised in other ways.
As you can see in the code snippet below, the text annotation is divided into categories: signs-symptom-mention, anatomical-site-mention and disease-disorder-mention.
xxxxxxxxxx
},
"disease-disorder-mention": {
"NEOPLASIA": [
"start: 2325",
"end: 2334",
"";polarity: 1",
"[codingScheme: SNOMEDCT_US, code: 1008369006, cui: C0027651, tui: T191]"
],
"CARCINOMA": [
"start: 3732",
"end: 3741",
"polarity: 1",
"[codingScheme: SNOMEDCT_US, code: 68453008, cui: C0007097, tui: T191J]"
],
},
"signs-symptom-mention": {
"TEST": [
"start: 95",
"end: 99",
"polarity: 1",
"[codingScheme:...]"
],
},
"anatomical-site-mention": {
"NIPPLE": [
"start: 2453",
"end: 2459",
"polarity: 1",
"[codingScheme:...]"
],
"UTERO": [
"start: 5652",
"end: 5657",
"polarity: 1",
"[codingScheme:...]"
],
"SKIN": [
"start: 2437",
"end: 2441",
"polarity: 1",
"[codingScheme:...]"
],
},
For each of these categories you have a key, this key is mapped to the entity that has been found in a medical dictionary — a list of all possible medical terms within a particular specialty. For each term you can find different objects: positional, start to end; polarity, representing the context in which a word has been found. A value of -1 when the word was found in a negative context; and 1 when the word was found in the affirmative. codingScheme is the medical dictionary where the match was found.
4. Model Ontology
The last step is modeling the ontology, and this part is really simple…
In order to map all the data together, coming in from SOPHIA’s NLP pipeline, you need to model the data against Grakn’s knowledge model.
Alessia provided [image below] screenshots from Grakn Workbase — Grakn’s IDE — visualizing her schema.
Breaking it down quickly and then we’ll look at a Graql code snippet of Alessia’s schema below. In the image above we can see that there are entities; guideline , which has thematic-sections, is further broken down into sentences. These entities are related to each other through a relation: guideline-contains-thematic-section, and thematic-section-contains-sentences.
The attributes are used to provide the specific pieces of rawtext: guideline-raw-text, thematic-section-raw-text, sentence-raw-text.
Continuing from the previous slide, sentence is connected to a token. This token matches the entity recognised by Apache cTAKES and is linked via a relation with the sub types of the entity medical-entity where each plays the role of mined-token. In this way we can see that Grakn uses type inheritance, allowing all sub types to inherit the attributes assigned to the parent entity: start, end, polarity, etc.
How Does This Look in Graql?
xxxxxxxxxx
define
medical-entity sub entity,
has start,
has end,
has polarity,
has [attribute-name],
plays minded-token;
token sub entity,
plays [role-name];
start sub attribute,
value: string;
end sub attribute,
value string;
polarity sub attribute,
value string;
ctakes-named-entity-recognition sub relation,
relates mined-token,
relates [role-name];
anatomical-site-mention sub medical-entity;
medication-mention sub medical-entity;
disease-disorder-mention sub medical-entity;
date-annotation sub medical-entity;
drug change status annotation sub medical-entity;
fraction-strength-annotation sub medical-entity;
measurement-annotation sub medical-entity;
strength-annotation sub medical-entity;
frequency-unit-annotation sub medical-entity;
sign-symptom-mention sub medical-entity;
The two other attributes defined in the schema that were highlighted in the presentation:
cui - concept unique identifier
This attribute assigns a concept code to medical-entity which is then shared among all the synonyms of the word.
tui - semantic concept unique identifier
This attribute is the semantic type of the word found.
SOPHIA in Practice
To put all of this into perspective, Alessia shared a hypothetical real world scenario where a doctor is able to make use of a CDSS.
Remember the problem:
How can a doctor provide a comprehensive and accurate recommendation to a patient, based on the doctor’s experience, patient history and up-to-date guidelines from the medical field; when they have, on average, 5 minutes in the room with the patient?
Meet Angela, a 45 year old patient with breast cancer. Angela goes to the doctor where she describes her symptoms (as in the slide above) and is examined. The doctor enters this information into the application, miFort, and goes through the signature approval steps before sending the report to SOPHIA as free text.
The report is then read and queried across Grakn to look for any matches. If such a match is found, a recommendation of action is provided to the doctor, with explanation of the links between the patient’s situation — represented by the medical report — and the knowledge base — represented by the guidelines or medical dictionaries. This is given as a bibliography for the doctor to reference.
Challenges Faced Along the Way
Ambiguous Entities
Ambiguous entities in the graph can bring about a host of other challenges; however, by reasoning not on the text itself but on the entity identification code — as shown above. Alessia never looks for a link between text but between concept codes (highlighted in red in the above slide). This ensures specificity and accuracy in her queries.
If you’re nervous about being precise or can’t find a code for what you need, here are 20 strange medical concept codes for coding diagnoses and symptoms.
A few highlights…
Space Adventures: V95.43 — spacecraft collision injuring occupant
Events Probably Not So Rare: W22.01 — walked into wall
That Should Never Happen: V97.33 — sucked into jet engine
Entities with the Same Meaning
This is relatively simple, as again, Alessia is reasoning not on the text itself but on the cui. Never looking for a link between text but between the cui attribute. So that when queried you are able to get the synonyms you want.
Concluding her talk, Alessia welcomed any contributions in the form of GraphQL integrations, self awareness of concepts within the knowledge graph or general interest in furthering the work of patient care through technology.
Special thank you to Alessia for her inspired work, contribution to the community and for always bringing joy into her work.
You can find the full presentation on the Grakn Labs YouTube channel here.
Opinions expressed by DZone contributors are their own.

Comments